Nuclear import of ribosomal
proteins: evidence for a novel type of nuclear localization signal
Rogier Stuger, Antonius C.J. Timmers, Hendrik
A. Raué, and Jan van 't Riet
in The ribosome: structure, function, antibiotics and cellular
interactions. RA Garrett, SR Douthwaite, A Liljas, AT Matheson, PB Moore,
HF Noller (eds). ASM Press, Washington DC (2000) pp 205-214
department of biochemistry
and molecular biology, Free University, Amsterdam
Abstract
The formation of eukaryotic
ribosomes is a highly complex process which requires the coordinated expression
of a large set of ribosomal genes, transcribed by three different RNA
polymerases, to ensure production of equimolar amounts of the four rRNAs
and the approximately 80 ribosomal protein (r-protein) species under all
growth conditions.
A
further level of complexity is added to eukaryotic ribosome biogenesis
by the fact that it involves different cellular compartments. Transcription
of the rRNA and r-protein genes take place in the nucleolus and nucleoplasm,
respectively. The r-protein mRNAs have to be exported to the cytoplasm.
After translation, the r-proteins must be imported into the nucle(ol)us,
where they have to be present in equimolar amounts to be assembled with
the rRNAs into ribosomal subunits. The subunits are then exported from
the nucleus to take up their function in the cytosol. Thus, ribosome biogenesis
in eukaryotic cells involves massive transport of macromolecules and macromolecular
complexes in both directions across the nuclear envelope. This transport
not only concerns the ribosomes themselves (or their components) but also
a large number of accessory factors, varying from constituents of the
transcription, translation, and splicing machinery to pre-rRNA-processing
and ribosome assembly factors.
In
recent years considerable insight has been gained into the mechanisms
of nucleocytoplasmic transport. Interestingly, a number of strong indications
were found that nuclear import of r-proteins uses a specialized import
pathway different from that used by the majority of karyophilic proteins.
This suggests that the nuclear localization signals or sequences (NLSs)
of r-proteins may be structurally distinct from the classical NLSs present
in the latter class of proteins. In this chapter we review present knowledge
of the mechanism and signals responsible for the nuclear import of proteins,
in particular r-proteins. A database search of the complete set of yeast
r-proteins for putative NLSs by using a set of criteria derived from the
comparison of all experimentally identified signals in yeast r-proteins
indicates that the large majority of these proteins may indeed possess
a novel type of NLS, characterized by the presence of a sequence motif
consisting of three basic residues within a 4- to 7-amino acid-long sequence,
of which the first, the last, or both are helix-breaking residues. [full-text
pdf] |
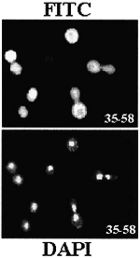 Nuclear
and nucleolar localization of Saccharomyces cerevisiae ribosomal
proteins S22 and S25
Nuclear
and nucleolar localization of Saccharomyces cerevisiae ribosomal
proteins S22 and S25